|
Nektar++
|
|
Nektar++
|
Cell model base class. More...
#include <CellModel.h>
Public Member Functions | |
| CellModel (const LibUtilities::SessionReaderSharedPtr &pSession, const MultiRegions::ExpListSharedPtr &pField) | |
| virtual | ~CellModel () |
| void | Initialise () |
| Initialise the cell model storage and set initial conditions. More... | |
| void | TimeIntegrate (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| Time integrate the cell model by one PDE timestep. More... | |
| void | Update (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| Compute the derivatives of cell model variables. More... | |
| void | GenerateSummary (SummaryList &s) |
| Print a summary of the cell model. More... | |
| unsigned int | GetNumCellVariables () |
| std::string | GetCellVarName (unsigned int idx) |
| Array< OneD, NekDouble > | GetCellSolutionCoeffs (unsigned int idx) |
| Array< OneD, NekDouble > | GetCellSolution (unsigned int idx) |
Protected Member Functions | |
| virtual void | v_Update (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time)=0 |
| virtual void | v_GenerateSummary (SummaryList &s)=0 |
| virtual std::string | v_GetCellVarName (unsigned int idx) |
| virtual void | v_SetInitialConditions ()=0 |
| void | LoadCellModel () |
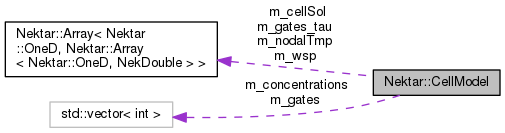
Protected Attributes | |
| LibUtilities::SessionReaderSharedPtr | m_session |
| Session. More... | |
| MultiRegions::ExpListSharedPtr | m_field |
| Transmembrane potential field from PDE system. More... | |
| int | m_nq |
| Number of physical points. More... | |
| int | m_nvar |
| Number of variables in cell model (inc. transmembrane voltage) More... | |
| NekDouble | m_lastTime |
| Timestep for pde model. More... | |
| int | m_substeps |
| Number of substeps to take. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_cellSol |
| Cell model solution variables. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_wsp |
| Cell model integration workspace. More... | |
| bool | m_useNodal |
| Flag indicating whether nodal projection in use. More... | |
| StdRegions::StdNodalTriExpSharedPtr | m_nodalTri |
| StdNodalTri for cell model calculations. More... | |
| StdRegions::StdNodalTetExpSharedPtr | m_nodalTet |
| Array< OneD, Array< OneD, NekDouble > > | m_nodalTmp |
| Temporary array for nodal projection. More... | |
| std::vector< int > | m_concentrations |
| Indices of cell model variables which are concentrations. More... | |
| std::vector< int > | m_gates |
| Indices of cell model variables which are gates. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_gates_tau |
| Storage for gate tau values. More... | |
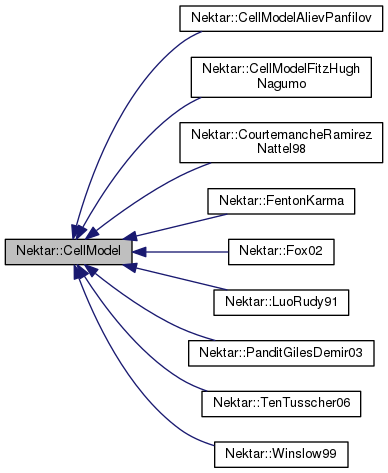
Cell model base class.
The CellModel class and derived classes implement a range of cell model ODE systems. A cell model comprises a system of ion concentration variables and zero or more gating variables. Gating variables are time-integrated using the Rush-Larsen method and for each variable y, the corresponding y_inf and tau_y value is computed by Update(). The tau values are stored in separate storage to inarray/outarray, m_gates_tau.
Definition at line 65 of file CellModel.h.
| Nektar::CellModel::CellModel | ( | const LibUtilities::SessionReaderSharedPtr & | pSession, |
| const MultiRegions::ExpListSharedPtr & | pField | ||
| ) |
Cell model base class constructor.
Definition at line 67 of file CellModel.cpp.
References Nektar::MemoryManager< DataType >::AllocateSharedPtr(), Nektar::LibUtilities::eGaussLobattoLegendre, Nektar::LibUtilities::eGaussRadauMAlpha1Beta0, Nektar::LibUtilities::eGaussRadauMAlpha2Beta0, Nektar::LibUtilities::eModified_A, Nektar::LibUtilities::eModified_B, Nektar::LibUtilities::eModified_C, Nektar::LibUtilities::eNodalTetEvenlySpaced, Nektar::LibUtilities::eNodalTriEvenlySpaced, Nektar::LibUtilities::eTetrahedron, Nektar::LibUtilities::eTriangle, m_field, m_lastTime, m_nodalTet, m_nodalTri, m_nq, m_nvar, m_session, m_substeps, and m_useNodal.
|
inlinevirtual |
Definition at line 71 of file CellModel.h.
|
inline |
Print a summary of the cell model.
Definition at line 92 of file CellModel.h.
References v_GenerateSummary().
Definition at line 320 of file CellModel.cpp.
References m_cellSol.
Definition at line 290 of file CellModel.cpp.
References ASSERTL0, Nektar::LibUtilities::eTriangle, m_cellSol, m_field, m_nodalTet, m_nodalTri, m_nvar, and m_useNodal.
|
inline |
Definition at line 102 of file CellModel.h.
References v_GetCellVarName().
Referenced by LoadCellModel().
|
inline |
Definition at line 97 of file CellModel.h.
References m_nvar.
| void Nektar::CellModel::Initialise | ( | ) |
Initialise the cell model storage and set initial conditions.
Initialise the cell model. Allocate workspace and variable storage.
Definition at line 126 of file CellModel.cpp.
References ASSERTL1, LoadCellModel(), m_cellSol, m_gates, m_gates_tau, m_nq, m_nvar, m_session, m_wsp, and v_SetInitialConditions().
|
protected |
Definition at line 325 of file CellModel.cpp.
References Nektar::MemoryManager< DataType >::AllocateSharedPtr(), Nektar::LibUtilities::eFunctionTypeExpression, Nektar::LibUtilities::eFunctionTypeFile, Nektar::LibUtilities::eTriangle, GetCellVarName(), Nektar::iterator, m_cellSol, m_field, m_lastTime, m_nodalTet, m_nodalTri, m_nq, m_session, m_useNodal, and Vmath::Zero().
Referenced by Initialise().
| void Nektar::CellModel::TimeIntegrate | ( | const Array< OneD, const Array< OneD, NekDouble > > & | inarray, |
| Array< OneD, Array< OneD, NekDouble > > & | outarray, | ||
| const NekDouble | time | ||
| ) |
Time integrate the cell model by one PDE timestep.
Integrates the cell model for one PDE time-step. Cell model is sub-stepped.
Ion concentrations and membrane potential are integrated using forward Euler, while gating variables are integrated using the Rush-Larsen scheme.
Definition at line 161 of file CellModel.cpp.
References Nektar::LibUtilities::eTriangle, m_cellSol, m_concentrations, m_field, m_gates, m_gates_tau, m_lastTime, m_nodalTet, m_nodalTmp, m_nodalTri, m_nq, m_substeps, m_useNodal, m_wsp, Vmath::Sdiv(), Vmath::Svtvp(), Update(), Vmath::Vcopy(), Vmath::Vexp(), Vmath::Vsub(), and Vmath::Vvtvp().
|
inline |
Compute the derivatives of cell model variables.
Definition at line 83 of file CellModel.h.
References v_Update().
Referenced by TimeIntegrate().
|
protectedpure virtual |
|
inlineprotectedvirtual |
Reimplemented in Nektar::CourtemancheRamirezNattel98, and Nektar::FentonKarma.
Definition at line 152 of file CellModel.h.
Referenced by GetCellVarName().
|
protectedpure virtual |
Implemented in Nektar::CourtemancheRamirezNattel98, Nektar::Winslow99, Nektar::FentonKarma, Nektar::LuoRudy91, Nektar::Fox02, Nektar::TenTusscher06, Nektar::CellModelFitzHughNagumo, and Nektar::CellModelAlievPanfilov.
Referenced by Initialise().
Cell model solution variables.
Definition at line 126 of file CellModel.h.
Referenced by GetCellSolution(), GetCellSolutionCoeffs(), Initialise(), LoadCellModel(), TimeIntegrate(), Nektar::CellModelAlievPanfilov::v_SetInitialConditions(), Nektar::CellModelFitzHughNagumo::v_SetInitialConditions(), Nektar::TenTusscher06::v_SetInitialConditions(), Nektar::Fox02::v_SetInitialConditions(), Nektar::LuoRudy91::v_SetInitialConditions(), Nektar::FentonKarma::v_SetInitialConditions(), Nektar::CourtemancheRamirezNattel98::v_SetInitialConditions(), and Nektar::Winslow99::v_SetInitialConditions().
|
protected |
Indices of cell model variables which are concentrations.
Definition at line 139 of file CellModel.h.
Referenced by Nektar::CellModelAlievPanfilov::CellModelAlievPanfilov(), Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo(), Nektar::CourtemancheRamirezNattel98::CourtemancheRamirezNattel98(), Nektar::Fox02::Fox02(), Nektar::LuoRudy91::LuoRudy91(), Nektar::TenTusscher06::TenTusscher06(), TimeIntegrate(), and Nektar::Winslow99::Winslow99().
|
protected |
Transmembrane potential field from PDE system.
Definition at line 115 of file CellModel.h.
Referenced by CellModel(), GetCellSolutionCoeffs(), LoadCellModel(), and TimeIntegrate().
|
protected |
Indices of cell model variables which are gates.
Definition at line 141 of file CellModel.h.
Referenced by Nektar::CourtemancheRamirezNattel98::CourtemancheRamirezNattel98(), Nektar::FentonKarma::FentonKarma(), Nektar::Fox02::Fox02(), Initialise(), Nektar::LuoRudy91::LuoRudy91(), Nektar::TenTusscher06::TenTusscher06(), TimeIntegrate(), and Nektar::Winslow99::Winslow99().
Storage for gate tau values.
Definition at line 143 of file CellModel.h.
Referenced by Initialise(), TimeIntegrate(), Nektar::TenTusscher06::v_Update(), Nektar::LuoRudy91::v_Update(), Nektar::FentonKarma::v_Update(), and Nektar::CourtemancheRamirezNattel98::v_Update().
|
protected |
Timestep for pde model.
Definition at line 121 of file CellModel.h.
Referenced by CellModel(), LoadCellModel(), and TimeIntegrate().
|
protected |
Definition at line 134 of file CellModel.h.
Referenced by CellModel(), GetCellSolutionCoeffs(), LoadCellModel(), and TimeIntegrate().
Temporary array for nodal projection.
Definition at line 136 of file CellModel.h.
Referenced by TimeIntegrate().
|
protected |
StdNodalTri for cell model calculations.
Definition at line 133 of file CellModel.h.
Referenced by CellModel(), GetCellSolutionCoeffs(), LoadCellModel(), and TimeIntegrate().
|
protected |
Number of physical points.
Definition at line 117 of file CellModel.h.
Referenced by CellModel(), Nektar::CellModelAlievPanfilov::CellModelAlievPanfilov(), Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo(), Initialise(), LoadCellModel(), Nektar::LuoRudy91::LuoRudy91(), Nektar::PanditGilesDemir03::PanditGilesDemir03(), TimeIntegrate(), Nektar::CellModelAlievPanfilov::v_SetInitialConditions(), Nektar::CellModelFitzHughNagumo::v_SetInitialConditions(), Nektar::Fox02::v_SetInitialConditions(), Nektar::TenTusscher06::v_SetInitialConditions(), Nektar::LuoRudy91::v_SetInitialConditions(), Nektar::FentonKarma::v_SetInitialConditions(), Nektar::Winslow99::v_SetInitialConditions(), Nektar::CourtemancheRamirezNattel98::v_SetInitialConditions(), Nektar::PanditGilesDemir03::v_Update(), Nektar::CellModelAlievPanfilov::v_Update(), Nektar::CellModelFitzHughNagumo::v_Update(), Nektar::TenTusscher06::v_Update(), Nektar::LuoRudy91::v_Update(), Nektar::Fox02::v_Update(), Nektar::Winslow99::v_Update(), Nektar::FentonKarma::v_Update(), and Nektar::CourtemancheRamirezNattel98::v_Update().
|
protected |
Number of variables in cell model (inc. transmembrane voltage)
Definition at line 119 of file CellModel.h.
Referenced by CellModel(), Nektar::CellModelAlievPanfilov::CellModelAlievPanfilov(), Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo(), Nektar::CourtemancheRamirezNattel98::CourtemancheRamirezNattel98(), Nektar::FentonKarma::FentonKarma(), Nektar::Fox02::Fox02(), GetCellSolutionCoeffs(), GetNumCellVariables(), Initialise(), Nektar::LuoRudy91::LuoRudy91(), Nektar::TenTusscher06::TenTusscher06(), and Nektar::Winslow99::Winslow99().
|
protected |
Session.
Definition at line 113 of file CellModel.h.
Referenced by CellModel(), Initialise(), and LoadCellModel().
|
protected |
Number of substeps to take.
Definition at line 123 of file CellModel.h.
Referenced by CellModel(), and TimeIntegrate().
|
protected |
Flag indicating whether nodal projection in use.
Definition at line 131 of file CellModel.h.
Referenced by CellModel(), GetCellSolutionCoeffs(), LoadCellModel(), and TimeIntegrate().
Cell model integration workspace.
Definition at line 128 of file CellModel.h.
Referenced by Initialise(), and TimeIntegrate().
 1.8.9.1
1.8.9.1