8.3 Session file configuration
8.3.1 Solver Info
Eqtype Specifies the PDE system to solve. The following values are supported:
Monodomain: solve the monodomain equation.
BidomainRoth: solve the bidomain equations using the Roth formulation.
CellModel Specifies the cell model to use. Available cell models are
| Value | Description | No. of Var. | Ref. |
AlievPanfilov | Phenomological | 1 | [2] |
CourtemancheRamirezNattel98 | Human atrial | 20 | [27] |
FitzHughNagumo | | | |
Fox02 | | | |
LuoRudy91 | Mammalian ventricular | 7 | [25] |
PanditGilesDemir03 | | | |
TenTusscher06 | Human ventricular | 18 | [45] |
Winslow99 | | | |
| |
Projection Specifies the Galerkin projection type to use. Only Continuous has been
extensively tested.
TimeIntegrationScheme Specifies the time integration scheme to use for advancing the
PDE system. This must be an IMEX scheme. Suitable choices are: IMEX Order 1,2,3,
IMEX, Variant dirk, Order 3, Free Parameters 3 4. The cell model state variables
are time advanced using Forward Euler for the ion concentrations, and Rush-Larsen for
the cell model gating variables.
DiffusionAdvancement Specifies whether the diffusion is handled implicitly
or explicitly in the time integration scheme. The current code only supports
Implicit integration of the diffusion term. The cell model is always integrated
explicitly.
8.3.2 Parameters
The following parameters can be specified in the PARAMETERS section of the session file.
Example values are taken from [11].
Chi sets the surface-to-volume ratio (Units: mm-1).
Example: χ = 140mm-1
Cm sets the specific membrane capacitance (Units: μFmm-2).
Example: Cm = 0.01μFmm-2
Substeps sets the number of substeps taken in time integrating the cell model for
each PDE timestep.
Example: 4
d_min, d_max, o_min, o_max specifies a bijective map to assign conductivity values σ to
intensity values μ when using the IsotropicConductivity function. The intensity map
is first thresholded to the range [dmin,dmax] and then the conductivity is calculated
as
σ = 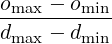 (1 - μ) + omin (1 - μ) + omin | | |
8.3.3 Functions
The following functions can be specified inside the CONDITIONS section of the session file. If
both are specified, the effect is multiplicative. Example values are taken from [11].
IsotropicConductivity specifies the conductivity σ of the tissue.
Example: σ = 0.13341 mSmm-1, based on σ = 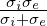 , σi = 0.17,σe =
0.62mSmm-1
, σi = 0.17,σe =
0.62mSmm-1
The variable name to use is intensity since the conductivity may be derived
from late-Gadolinium enhanced MRA imaging. Example specifications are
1<E VAR="intensity" VALUE="0.13341" />
2<F VAR="intensity" FILE="scarmap.con" />
where scarmap.con is a Nektar++ field file containing a variable intensity describing
the conductivity across the domain.
AnisotropicConductivity specifies the conductivity σ of the tissue.
8.3.4 Filters
The following filters are supported exclusively for the cardiac EP solver. Further filters from
section 3.4 are also available for this solver.
Benchmark (section 3.4.3)
CellHistoryPoints (section 3.4.4)
CheckpointCellModel (section 3.4.5)
Electrogram (section 3.4.7)
8.3.5 Stimuli
Electrophysiological propagaion is initiated through the stimulus current Iion. The STIMULI
section describes one or more regions of stimulus and the time-dependent protocol with which
they are applied.
1<STIMULI>
2 ...
3</STIMULI>
A number of stimulus types are available
8.3.5.1 Stimulus types
StimulusRect stimulates a cuboid-shaped region of the domain, specified by two
coordinates (x1,y1,z1) and (x2,y2,z2). An additional parameter specifies the
"smoothness" of the boundaries of the region; higher values produce a sharper boundary.
Finally, the maximum strength of the stimulus current is specified in μA∕mm3
1<STIMULUS TYPE="StimulusRect" ID="0">
2 <p_x1> -15.24 </p_x1>
3 <p_y1> 14.02 </p_y1>
4 <p_z1> 6.87 </p_z1>
5 <p_x2> 12.23 </p_x2>
6 <p_y2> 16.56 </p_y2>
7 <p_z2> 8.88 </p_z2>
8 <p_is> 100.00 </p_is>
9 <p_strength> 50.0 </p_strength>
10</STIMULUS>
StimulusCirc stimulates a spherical region of the domain, as specified by a centre and
radius. The smoothness and strength parameters are also specified as for ‘StimulusRect‘.
1<STIMULUS TYPE="StimulusCirc" ID="0">
2 <p_x1> -15.24 </p_x1>
3 <p_y1> 14.02 </p_y1>
4 <p_z1> 6.87 </p_z1>
5 <p_r1> 12.23 </p_r1>
6 <p_is> 100.00 </p_is>
7 <p_strength> 50.0 </p_strength>
8</STIMULUS>
8.3.5.2 Protocols
A protocol specifies the time-dependent function indicating the strength of the stimulus and
one such PROTOCOL section should be included within each STIMULUS. This can be expressed as
one of:
ProtocolSingle a single stimulus is applied at a given start time and for a given
duration
1<PROTOCOL TYPE="ProtocolSingle">
2 <START> 0.0 </START>
3 <DURATION> 2.0 </DURATION>
4</PROTOCOL>
ProtocolS1 a train of pulses of fixed duration applied at a given start time and with a
given cycle length.
1<PROTOCOL TYPE="ProtocolS1">
2 <START> 0.0 </START>
3 <DURATION> 2.0 </DURATION>
4 <S1CYCLELENGTH> 300.0 </S1CYCLELENGTH>
5 <NUM_S1> 5 </NUM_S1>
6</PROTOCOL>
ProtocolS1S2 same as ‘ProtocolS1‘ except with an additional single pulse applied at a
different cycle length at the end of the train of S1 pulses.
1<PROTOCOL TYPE="ProtocolS1S2">
2 <START> 0.0 </START>
3 <DURATION> 2.0 </DURATION>
4 <S1CYCLELENGTH> 300.0 </S1CYCLELENGTH>
5 <NUM_S1> 5 </NUM_S1>
6 <S2CYCLELENGTH> 100.0 </S2CYCLELENGTH>
7</PROTOCOL>
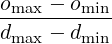 (1 - μ) + omin
(1 - μ) + omin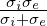 , σi = 0.17,σe =
0.62mSmm-1
, σi = 0.17,σe =
0.62mSmm-1