|
Nektar++
|
|
Nektar++
|
Cell model base class. More...
#include <CellModel.h>
Public Member Functions | |
| CellModel (const LibUtilities::SessionReaderSharedPtr &pSession, const MultiRegions::ExpListSharedPtr &pField) | |
| virtual | ~CellModel () |
| void | Initialise () |
| Initialise the cell model storage and set initial conditions. More... | |
| void | TimeIntegrate (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| Time integrate the cell model by one PDE timestep. More... | |
| void | Update (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| Compute the derivatives of cell model variables. More... | |
| void | GenerateSummary (SummaryList &s) |
| Print a summary of the cell model. More... | |
| unsigned int | GetNumCellVariables () |
| std::string | GetCellVarName (unsigned int idx) |
| Array< OneD, NekDouble > | GetCellSolutionCoeffs (unsigned int idx) |
| Array< OneD, NekDouble > | GetCellSolution (unsigned int idx) |
Protected Member Functions | |
| virtual void | v_Update (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time)=0 |
| virtual void | v_GenerateSummary (SummaryList &s)=0 |
| virtual std::string | v_GetCellVarName (unsigned int idx) |
| virtual void | v_SetInitialConditions ()=0 |
| void | LoadCellModel () |
Protected Attributes | |
| LibUtilities::SessionReaderSharedPtr | m_session |
| Session. More... | |
| MultiRegions::ExpListSharedPtr | m_field |
| Transmembrane potential field from PDE system. More... | |
| int | m_nq |
| Number of physical points. More... | |
| int | m_nvar |
| Number of variables in cell model (inc. transmembrane voltage) More... | |
| NekDouble | m_lastTime |
| Timestep for pde model. More... | |
| int | m_substeps |
| Number of substeps to take. More... | |
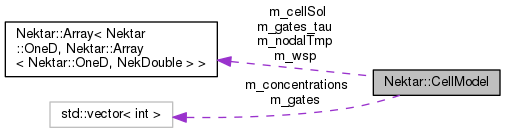
| Array< OneD, Array< OneD, NekDouble > > | m_cellSol |
| Cell model solution variables. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_wsp |
| Cell model integration workspace. More... | |
| bool | m_useNodal |
| Flag indicating whether nodal projection in use. More... | |
| StdRegions::StdNodalTriExpSharedPtr | m_nodalTri |
| StdNodalTri for cell model calculations. More... | |
| StdRegions::StdNodalTetExpSharedPtr | m_nodalTet |
| Array< OneD, Array< OneD, NekDouble > > | m_nodalTmp |
| Temporary array for nodal projection. More... | |
| std::vector< int > | m_concentrations |
| Indices of cell model variables which are concentrations. More... | |
| std::vector< int > | m_gates |
| Indices of cell model variables which are gates. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_gates_tau |
| Storage for gate tau values. More... | |
Cell model base class.
The CellModel class and derived classes implement a range of cell model ODE systems. A cell model comprises a system of ion concentration variables and zero or more gating variables. Gating variables are time-integrated using the Rush-Larsen method and for each variable y, the corresponding y_inf and tau_y value is computed by Update(). The tau values are stored in separate storage to inarray/outarray, m_gates_tau.
Definition at line 65 of file CellModel.h.
| Nektar::CellModel::CellModel | ( | const LibUtilities::SessionReaderSharedPtr & | pSession, |
| const MultiRegions::ExpListSharedPtr & | pField | ||
| ) |
Cell model base class constructor.
Definition at line 69 of file CellModel.cpp.
References Nektar::LibUtilities::eGaussLobattoLegendre, Nektar::LibUtilities::eGaussRadauMAlpha1Beta0, Nektar::LibUtilities::eGaussRadauMAlpha2Beta0, Nektar::LibUtilities::eModified_A, Nektar::LibUtilities::eModified_B, Nektar::LibUtilities::eModified_C, Nektar::LibUtilities::eNodalTetEvenlySpaced, Nektar::LibUtilities::eNodalTriEvenlySpaced, Nektar::LibUtilities::eTetrahedron, and Nektar::LibUtilities::eTriangle.
|
inlinevirtual |
Definition at line 71 of file CellModel.h.
|
inline |
Print a summary of the cell model.
Definition at line 92 of file CellModel.h.
References v_GenerateSummary().
Definition at line 322 of file CellModel.cpp.
Definition at line 292 of file CellModel.cpp.
References ASSERTL0, and Nektar::LibUtilities::eTriangle.
|
inline |
Definition at line 102 of file CellModel.h.
References v_GetCellVarName().
|
inline |
Definition at line 97 of file CellModel.h.
References m_nvar.
| void Nektar::CellModel::Initialise | ( | ) |
Initialise the cell model storage and set initial conditions.
Initialise the cell model. Allocate workspace and variable storage.
Definition at line 128 of file CellModel.cpp.
References ASSERTL1.
|
protected |
Definition at line 327 of file CellModel.cpp.
References Nektar::LibUtilities::eFunctionTypeExpression, Nektar::LibUtilities::eFunctionTypeFile, Nektar::LibUtilities::eTriangle, Nektar::iterator, and Vmath::Zero().
| void Nektar::CellModel::TimeIntegrate | ( | const Array< OneD, const Array< OneD, NekDouble > > & | inarray, |
| Array< OneD, Array< OneD, NekDouble > > & | outarray, | ||
| const NekDouble | time | ||
| ) |
Time integrate the cell model by one PDE timestep.
Integrates the cell model for one PDE time-step. Cell model is sub-stepped.
Ion concentrations and membrane potential are integrated using forward Euler, while gating variables are integrated using the Rush-Larsen scheme.
Definition at line 163 of file CellModel.cpp.
References Nektar::LibUtilities::eTriangle, Vmath::Sdiv(), Vmath::Svtvp(), Vmath::Vcopy(), Vmath::Vexp(), Vmath::Vsub(), and Vmath::Vvtvp().
|
inline |
Compute the derivatives of cell model variables.
Definition at line 83 of file CellModel.h.
References v_Update().
|
protectedpure virtual |
|
inlineprotectedvirtual |
Reimplemented in Nektar::CourtemancheRamirezNattel98, and Nektar::FentonKarma.
Definition at line 152 of file CellModel.h.
Referenced by GetCellVarName().
|
protectedpure virtual |
Cell model solution variables.
Definition at line 126 of file CellModel.h.
Referenced by Nektar::CellModelAlievPanfilov::v_SetInitialConditions(), Nektar::CellModelFitzHughNagumo::v_SetInitialConditions(), Nektar::Fox02::v_SetInitialConditions(), Nektar::TenTusscher06::v_SetInitialConditions(), Nektar::LuoRudy91::v_SetInitialConditions(), Nektar::FentonKarma::v_SetInitialConditions(), Nektar::CourtemancheRamirezNattel98::v_SetInitialConditions(), and Nektar::Winslow99::v_SetInitialConditions().
|
protected |
Indices of cell model variables which are concentrations.
Definition at line 139 of file CellModel.h.
Referenced by Nektar::CellModelAlievPanfilov::CellModelAlievPanfilov(), Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo(), Nektar::CourtemancheRamirezNattel98::CourtemancheRamirezNattel98(), Nektar::Fox02::Fox02(), Nektar::LuoRudy91::LuoRudy91(), Nektar::TenTusscher06::TenTusscher06(), and Nektar::Winslow99::Winslow99().
|
protected |
Transmembrane potential field from PDE system.
Definition at line 115 of file CellModel.h.
|
protected |
Indices of cell model variables which are gates.
Definition at line 141 of file CellModel.h.
Referenced by Nektar::CourtemancheRamirezNattel98::CourtemancheRamirezNattel98(), Nektar::FentonKarma::FentonKarma(), Nektar::Fox02::Fox02(), Nektar::LuoRudy91::LuoRudy91(), Nektar::TenTusscher06::TenTusscher06(), and Nektar::Winslow99::Winslow99().
Storage for gate tau values.
Definition at line 143 of file CellModel.h.
Referenced by Nektar::TenTusscher06::v_Update(), Nektar::LuoRudy91::v_Update(), Nektar::FentonKarma::v_Update(), and Nektar::CourtemancheRamirezNattel98::v_Update().
|
protected |
Timestep for pde model.
Definition at line 121 of file CellModel.h.
|
protected |
Definition at line 134 of file CellModel.h.
Temporary array for nodal projection.
Definition at line 136 of file CellModel.h.
|
protected |
StdNodalTri for cell model calculations.
Definition at line 133 of file CellModel.h.
|
protected |
Number of physical points.
Definition at line 117 of file CellModel.h.
Referenced by Nektar::CellModelAlievPanfilov::CellModelAlievPanfilov(), Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo(), Nektar::LuoRudy91::LuoRudy91(), Nektar::PanditGilesDemir03::PanditGilesDemir03(), Nektar::CellModelAlievPanfilov::v_SetInitialConditions(), Nektar::CellModelFitzHughNagumo::v_SetInitialConditions(), Nektar::TenTusscher06::v_SetInitialConditions(), Nektar::Fox02::v_SetInitialConditions(), Nektar::LuoRudy91::v_SetInitialConditions(), Nektar::FentonKarma::v_SetInitialConditions(), Nektar::CourtemancheRamirezNattel98::v_SetInitialConditions(), Nektar::Winslow99::v_SetInitialConditions(), Nektar::PanditGilesDemir03::v_Update(), Nektar::CellModelAlievPanfilov::v_Update(), Nektar::CellModelFitzHughNagumo::v_Update(), Nektar::TenTusscher06::v_Update(), Nektar::Fox02::v_Update(), Nektar::LuoRudy91::v_Update(), Nektar::Winslow99::v_Update(), Nektar::FentonKarma::v_Update(), and Nektar::CourtemancheRamirezNattel98::v_Update().
|
protected |
Number of variables in cell model (inc. transmembrane voltage)
Definition at line 119 of file CellModel.h.
Referenced by Nektar::CellModelAlievPanfilov::CellModelAlievPanfilov(), Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo(), Nektar::CourtemancheRamirezNattel98::CourtemancheRamirezNattel98(), Nektar::FentonKarma::FentonKarma(), Nektar::Fox02::Fox02(), GetNumCellVariables(), Nektar::LuoRudy91::LuoRudy91(), Nektar::TenTusscher06::TenTusscher06(), and Nektar::Winslow99::Winslow99().
|
protected |
Session.
Definition at line 113 of file CellModel.h.
|
protected |
Number of substeps to take.
Definition at line 123 of file CellModel.h.
|
protected |
Flag indicating whether nodal projection in use.
Definition at line 131 of file CellModel.h.
 1.8.8
1.8.8