|
Nektar++
|
|
Nektar++
|
FitzHugh-Nagumo model. More...
#include <FitzhughNagumo.h>
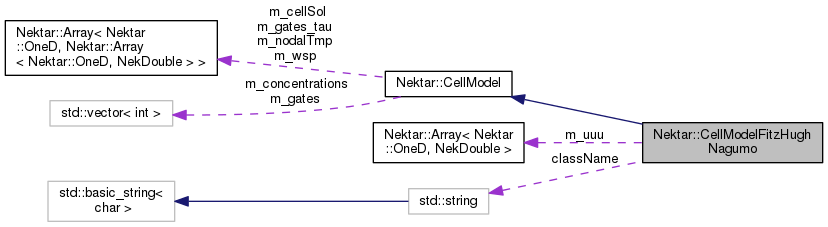
Public Member Functions | |
| CellModelFitzHughNagumo (const LibUtilities::SessionReaderSharedPtr &pSession, const MultiRegions::ExpListSharedPtr &pField) | |
| virtual | ~CellModelFitzHughNagumo () |
 Public Member Functions inherited from Nektar::CellModel Public Member Functions inherited from Nektar::CellModel | |
| CellModel (const LibUtilities::SessionReaderSharedPtr &pSession, const MultiRegions::ExpListSharedPtr &pField) | |
| virtual | ~CellModel () |
| void | Initialise () |
| Initialise the cell model storage and set initial conditions. More... | |
| void | TimeIntegrate (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| Time integrate the cell model by one PDE timestep. More... | |
| void | Update (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| Compute the derivatives of cell model variables. More... | |
| void | GenerateSummary (SummaryList &s) |
| Print a summary of the cell model. More... | |
| unsigned int | GetNumCellVariables () |
| std::string | GetCellVarName (unsigned int idx) |
| Array< OneD, NekDouble > | GetCellSolutionCoeffs (unsigned int idx) |
| Array< OneD, NekDouble > | GetCellSolution (unsigned int idx) |
Static Public Member Functions | |
| static CellModelSharedPtr | create (const LibUtilities::SessionReaderSharedPtr &pSession, const MultiRegions::ExpListSharedPtr &pField) |
| Creates an instance of this class. More... | |
Static Public Attributes | |
| static std::string | className |
| Name of class. More... | |
Protected Member Functions | |
| virtual void | v_Update (const Array< OneD, const Array< OneD, NekDouble > > &inarray, Array< OneD, Array< OneD, NekDouble > > &outarray, const NekDouble time) |
| virtual void | v_GenerateSummary (SummaryList &s) |
| virtual void | v_SetInitialConditions () |
 Protected Member Functions inherited from Nektar::CellModel Protected Member Functions inherited from Nektar::CellModel | |
| virtual std::string | v_GetCellVarName (unsigned int idx) |
| void | LoadCellModel () |
Private Attributes | |
| NekDouble | m_beta |
| NekDouble | m_epsilon |
| Array< OneD, NekDouble > | m_uuu |
Temporary space for storing 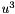 when computing reaction term. More... when computing reaction term. More... | |
Additional Inherited Members | |
 Protected Attributes inherited from Nektar::CellModel Protected Attributes inherited from Nektar::CellModel | |
| LibUtilities::SessionReaderSharedPtr | m_session |
| Session. More... | |
| MultiRegions::ExpListSharedPtr | m_field |
| Transmembrane potential field from PDE system. More... | |
| int | m_nq |
| Number of physical points. More... | |
| int | m_nvar |
| Number of variables in cell model (inc. transmembrane voltage) More... | |
| NekDouble | m_lastTime |
| Timestep for pde model. More... | |
| int | m_substeps |
| Number of substeps to take. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_cellSol |
| Cell model solution variables. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_wsp |
| Cell model integration workspace. More... | |
| bool | m_useNodal |
| Flag indicating whether nodal projection in use. More... | |
| StdRegions::StdNodalTriExpSharedPtr | m_nodalTri |
| StdNodalTri for cell model calculations. More... | |
| StdRegions::StdNodalTetExpSharedPtr | m_nodalTet |
| Array< OneD, Array< OneD, NekDouble > > | m_nodalTmp |
| Temporary array for nodal projection. More... | |
| std::vector< int > | m_concentrations |
| Indices of cell model variables which are concentrations. More... | |
| std::vector< int > | m_gates |
| Indices of cell model variables which are gates. More... | |
| Array< OneD, Array< OneD, NekDouble > > | m_gates_tau |
| Storage for gate tau values. More... | |
FitzHugh-Nagumo model.
Definition at line 44 of file FitzhughNagumo.h.
| Nektar::CellModelFitzHughNagumo::CellModelFitzHughNagumo | ( | const LibUtilities::SessionReaderSharedPtr & | pSession, |
| const MultiRegions::ExpListSharedPtr & | pField | ||
| ) |
Definition at line 53 of file FitzhughNagumo.cpp.
References m_beta, Nektar::CellModel::m_concentrations, m_epsilon, Nektar::CellModel::m_nq, Nektar::CellModel::m_nvar, and m_uuu.
|
inlinevirtual |
Definition at line 62 of file FitzhughNagumo.h.
|
inlinestatic |
Creates an instance of this class.
Definition at line 48 of file FitzhughNagumo.h.
References Nektar::MemoryManager< DataType >::AllocateSharedPtr().
|
protectedvirtual |
Implements Nektar::CellModel.
Definition at line 97 of file FitzhughNagumo.cpp.
References Nektar::SolverUtils::AddSummaryItem(), and m_beta.
|
protectedvirtual |
Implements Nektar::CellModel.
Definition at line 107 of file FitzhughNagumo.cpp.
References Vmath::Fill(), Nektar::CellModel::m_cellSol, and Nektar::CellModel::m_nq.
|
protectedvirtual |
Implements Nektar::CellModel.
Definition at line 68 of file FitzhughNagumo.cpp.
References m_beta, m_epsilon, Nektar::CellModel::m_nq, m_uuu, Vmath::Sadd(), Vmath::Smul(), Vmath::Svtvp(), Vmath::Vmul(), and Vmath::Vsub().
|
static |
Name of class.
Registers the class with the Factory.
Definition at line 56 of file FitzhughNagumo.h.
|
private |
Definition at line 75 of file FitzhughNagumo.h.
Referenced by CellModelFitzHughNagumo(), v_GenerateSummary(), and v_Update().
|
private |
Definition at line 76 of file FitzhughNagumo.h.
Referenced by CellModelFitzHughNagumo(), and v_Update().
Temporary space for storing 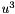 when computing reaction term.
when computing reaction term.
Definition at line 79 of file FitzhughNagumo.h.
Referenced by CellModelFitzHughNagumo(), and v_Update().
 1.8.8
1.8.8