|
Nektar++
|
|
Nektar++
|
#include <PreconditionerLowEnergy.h>
Static Public Member Functions | |
| static PreconditionerSharedPtr | create (const boost::shared_ptr< GlobalLinSys > &plinsys, const boost::shared_ptr< AssemblyMap > &pLocToGloMap) |
| Creates an instance of this class. | |
Static Public Attributes | |
| static std::string | className |
| Name of class. | |
Private Member Functions | |
| void | SetUpReferenceElements (void) |
| Sets up the reference elements needed by the preconditioner. | |
| void | CreateMultiplicityMap (void) |
| void | SetupBlockTransformationMatrix (void) |
| void | ModifyPrismTransformationMatrix (LocalRegions::TetExpSharedPtr TetExp, LocalRegions::PrismExpSharedPtr PrismExp, DNekMatSharedPtr Rmodprism, DNekMatSharedPtr RTmodprism) |
| Modify the prism transformation matrix to align with the tetrahedral modes. | |
| SpatialDomains::TetGeomSharedPtr | CreateRefTetGeom (void) |
| Sets up the reference tretrahedral element needed to construct *a low energy basis. | |
| SpatialDomains::PrismGeomSharedPtr | CreateRefPrismGeom (void) |
| Sets up the reference prismatic element needed to construct *a low energy basis. | |
| SpatialDomains::HexGeomSharedPtr | CreateRefHexGeom (void) |
| Sets up the reference hexahedral element needed to construct *a low energy basis. | |
| virtual void | v_InitObject () |
| virtual void | v_DoPreconditioner (const Array< OneD, NekDouble > &pInput, Array< OneD, NekDouble > &pOutput) |
| virtual void | v_DoTransformToLowEnergy (Array< OneD, NekDouble > &pInOut, int offset) |
| Transform the solution vector vector to low energy. | |
| virtual void | v_DoTransformToLowEnergy (const Array< OneD, NekDouble > &pInput, Array< OneD, NekDouble > &pOutput) |
| Transform the solution vector vector to low energy. | |
| virtual void | v_DoTransformFromLowEnergy (Array< OneD, NekDouble > &pInOut) |
| transform the solution vector from low energy back to the original basis. | |
| virtual void | v_DoMultiplybyInverseTransformationMatrix (const Array< OneD, NekDouble > &pInput, Array< OneD, NekDouble > &pOutput) |
| Multiply by the block inverse transformation matrix. | |
| virtual void | v_DoMultiplybyInverseTransposedTransformationMatrix (const Array< OneD, NekDouble > &pInput, Array< OneD, NekDouble > &pOutput) |
| Multiply by the block tranposed inverse transformation matrix. | |
| virtual void | v_BuildPreconditioner () |
Construct the low energy preconditioner from 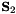 . . | |
| virtual DNekScalMatSharedPtr | v_TransformedSchurCompl (int offset, const boost::shared_ptr< DNekScalMat > &loc_mat) |
| Set up the transformed block matrix system. | |
Additional Inherited Members | |
 Protected Member Functions inherited from Nektar::MultiRegions::Preconditioner Protected Member Functions inherited from Nektar::MultiRegions::Preconditioner | |
| virtual DNekScalMatSharedPtr | v_TransformedSchurCompl (int offset, const boost::shared_ptr< DNekScalMat > &loc_mat) |
Get block elemental transposed transformation matrix 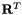 . . | |
This class implements low energy preconditioning for the conjugate
gradient matrix solver.
Definition at line 53 of file PreconditionerLowEnergy.h.
| Nektar::MultiRegions::PreconditionerLowEnergy::PreconditionerLowEnergy | ( | const boost::shared_ptr< GlobalLinSys > & | plinsys, |
| const AssemblyMapSharedPtr & | pLocToGloMap | ||
| ) |
Definition at line 64 of file PreconditionerLowEnergy.cpp.
|
inlinevirtual |
Definition at line 75 of file PreconditionerLowEnergy.h.
|
inlinestatic |
Creates an instance of this class.
Definition at line 57 of file PreconditionerLowEnergy.h.
Create the inverse multiplicity map.
Definition at line 1271 of file PreconditionerLowEnergy.cpp.
References m_locToGloMap, m_locToGloSignMult, m_multiplicity, Vmath::Sdiv(), and sign.
Referenced by v_InitObject().
|
private |
Sets up the reference hexahedral element needed to construct *a low energy basis.
Definition at line 1510 of file PreconditionerLowEnergy.cpp.
References Nektar::StdRegions::eBackwards, and Nektar::StdRegions::eForwards.
Referenced by SetUpReferenceElements().
|
private |
Sets up the reference prismatic element needed to construct *a low energy basis.
Definition at line 1328 of file PreconditionerLowEnergy.cpp.
References Nektar::StdRegions::eBackwards, and Nektar::StdRegions::eForwards.
Referenced by SetUpReferenceElements().
|
private |
Sets up the reference tretrahedral element needed to construct *a low energy basis.
Definition at line 1418 of file PreconditionerLowEnergy.cpp.
References Nektar::StdRegions::eBackwards, and Nektar::StdRegions::eForwards.
Referenced by SetUpReferenceElements().
|
private |
Modify the prism transformation matrix to align with the tetrahedral modes.
This routine replaces the edge and triangular face components of the prismatic vertex transformation matrices 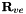 and
and 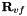 with the corresponding components from the tetrahedral transformation matrices. Additionally, triangular face components in the prismatic edge transformation matrix
with the corresponding components from the tetrahedral transformation matrices. Additionally, triangular face components in the prismatic edge transformation matrix 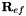 with the corresponding component from the tetrahedral transformation matrix.
with the corresponding component from the tetrahedral transformation matrix.
Definition at line 1867 of file PreconditionerLowEnergy.cpp.
Referenced by SetUpReferenceElements().
Set a block transformation matrices for each element type. These are needed in routines that transform the schur complement matrix to and from the low energy basis.
Definition at line 912 of file PreconditionerLowEnergy.cpp.
References Nektar::eDIAGONAL, Nektar::LibUtilities::eHexahedron, Nektar::LibUtilities::ePrism, Nektar::LibUtilities::eTetrahedron, m_InvRBlk, m_InvRTBlk, m_linsys, m_locToGloMap, m_RBlk, m_Rhex, m_Rinvhex, m_Rinvprism, m_Rinvtet, m_Rprism, m_RTBlk, m_Rtet, m_RThex, m_RTinvhex, m_RTinvprism, m_RTinvtet, m_RTprism, and m_RTtet.
Referenced by v_InitObject().
Sets up the reference elements needed by the preconditioner.
Sets up reference elements which are used to preconditioning the corresponding matrices. Currently we support tetrahedral, prismatic and hexahedral elements
Definition at line 1599 of file PreconditionerLowEnergy.cpp.
References CreateRefHexGeom(), CreateRefPrismGeom(), CreateRefTetGeom(), Nektar::eFULL, Nektar::LibUtilities::eGaussLobattoLegendre, Nektar::LibUtilities::eGaussRadauMAlpha1Beta0, Nektar::LibUtilities::eGaussRadauMAlpha2Beta0, Nektar::LibUtilities::eHexahedron, Nektar::StdRegions::eMass, Nektar::LibUtilities::eModified_A, Nektar::LibUtilities::eModified_B, Nektar::LibUtilities::eModified_C, Nektar::StdRegions::ePreconR, Nektar::StdRegions::ePreconRMass, Nektar::StdRegions::ePreconRT, Nektar::StdRegions::ePreconRTMass, Nektar::LibUtilities::ePrism, Nektar::LibUtilities::eTetrahedron, Nektar::MultiRegions::GlobalMatrixKey::GetConstFactors(), Nektar::MultiRegions::GlobalMatrixKey::GetMatrixType(), Nektar::MultiRegions::GlobalMatrixKey::GetNVarCoeffs(), Nektar::MultiRegions::GlobalMatrixKey::GetVarCoeffs(), m_linsys, m_Rhex, m_Rinvhex, m_Rinvprism, m_Rinvtet, m_Rprism, m_Rtet, m_RThex, m_RTinvhex, m_RTinvprism, m_RTinvtet, m_RTprism, m_RTtet, and ModifyPrismTransformationMatrix().
Referenced by v_InitObject().
|
privatevirtual |
Construct the low energy preconditioner from 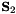 .
.
![\[\mathbf{M}^{-1}=\left[\begin{array}{ccc} Diag[(\mathbf{S_{2}})_{vv}] & & \\ & (\mathbf{S}_{2})_{eb} & \\ & & (\mathbf{S}_{2})_{fb} \end{array}\right] \]](form_402.png)
where 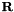 is the transformation matrix and
is the transformation matrix and 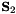 the Schur complement of the modified basis, given by
the Schur complement of the modified basis, given by
![\[\mathbf{S}_{2}=\mathbf{R}\mathbf{S}_{1}\mathbf{R}^{T}\]](form_403.png)
where 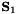 is the local schur complement matrix for each element.
is the local schur complement matrix for each element.
Definition at line 118 of file PreconditionerLowEnergy.cpp.
References Nektar::StdRegions::StdExpansion::as(), Vmath::Assmb(), Nektar::MultiRegions::DeterminePeriodicFaceOrient(), Nektar::eDIAGONAL, Nektar::SpatialDomains::eDirichlet, Nektar::eFULL, Nektar::LibUtilities::eHexahedron, Nektar::LibUtilities::ePrism, Nektar::LibUtilities::eTetrahedron, Gs::Gather(), Nektar::StdRegions::StdExpansion::GetEdgeInverseBoundaryMap(), Nektar::StdRegions::StdExpansion::GetEdgeNcoeffs(), Nektar::StdRegions::StdExpansion::GetFaceIntNcoeffs(), Nektar::StdRegions::StdExpansion::GetForient(), Gs::gs_add, Gs::Init(), m_BlkMat, Nektar::MultiRegions::Preconditioner::m_comm, m_linsys, m_locToGloMap, m_RBlk, m_Rhex, m_Rprism, m_RTBlk, m_Rtet, m_RThex, m_RTprism, m_RTtet, and Nektar::LibUtilities::ReduceMax.
|
privatevirtual |
Multiply by the block inverse transformation matrix.
Definition at line 1127 of file PreconditionerLowEnergy.cpp.
References ASSERTL1, Nektar::eWrapper, Vmath::Gathr(), m_InvRBlk, m_locToGloMap, m_locToGloSignMult, m_map, and Vmath::Vcopy().
|
privatevirtual |
Multiply by the block tranposed inverse transformation matrix.
Definition at line 1172 of file PreconditionerLowEnergy.cpp.
References ASSERTL1, Nektar::eWrapper, m_InvRTBlk, m_locToGloMap, m_map, m_multiplicity, and Vmath::Vmul().
|
privatevirtual |
Apply the low energy preconditioner during the conjugate gradient routine
Definition at line 891 of file PreconditionerLowEnergy.cpp.
References Nektar::eWrapper, and m_locToGloMap.
|
privatevirtual |
transform the solution vector from low energy back to the original basis.
After the conjugate gradient routine the output vector is in the low energy basis and must be trasnformed back to the original basis in order to get the correct solution out. the solution vector i.e. 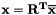 .
.
Definition at line 1085 of file PreconditionerLowEnergy.cpp.
References ASSERTL1, Vmath::Assmb(), Nektar::eWrapper, m_locToGloMap, m_locToGloSignMult, m_map, m_RTBlk, and Vmath::Vcopy().
|
privatevirtual |
Transform the solution vector vector to low energy.
As the conjugate gradient system is solved for the low energy basis, the solution vector 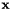 must be transformed to the low energy basis i.e.
must be transformed to the low energy basis i.e. 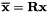 .
.
Definition at line 993 of file PreconditionerLowEnergy.cpp.
References Nektar::eWrapper, Vmath::Gathr(), m_locToGloMap, m_locToGloSignMult, m_map, m_RBlk, and Vmath::Vcopy().
|
privatevirtual |
Transform the solution vector vector to low energy.
As the conjugate gradient system is solved for the low energy basis, the solution vector 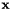 must be transformed to the low energy basis i.e.
must be transformed to the low energy basis i.e. 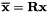 .
.
Definition at line 1034 of file PreconditionerLowEnergy.cpp.
References ASSERTL1, Nektar::eWrapper, Vmath::Gathr(), m_locToGloMap, m_locToGloSignMult, m_map, m_RBlk, and Vmath::Vcopy().
|
privatevirtual |
Definition at line 73 of file PreconditionerLowEnergy.cpp.
References ASSERTL0, CreateMultiplicityMap(), Nektar::MultiRegions::eIterativeStaticCond, m_linsys, m_locToGloMap, SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
privatevirtual |
Set up the transformed block matrix system.
Sets up a block elemental matrix in which each of the block matrix is the low energy equivalent i.e. 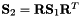
Definition at line 1216 of file PreconditionerLowEnergy.cpp.
References Nektar::eFULL, Nektar::LibUtilities::eHexahedron, Nektar::LibUtilities::ePrism, Nektar::LibUtilities::eTetrahedron, m_linsys, m_Rhex, m_Rprism, m_Rtet, m_RThex, m_RTprism, and m_RTtet.
|
static |
Name of class.
Registers the class with the Factory.
Definition at line 68 of file PreconditionerLowEnergy.h.
|
protected |
Definition at line 82 of file PreconditionerLowEnergy.h.
Referenced by v_BuildPreconditioner().
|
protected |
Definition at line 83 of file PreconditionerLowEnergy.h.
|
protected |
Definition at line 87 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and v_DoMultiplybyInverseTransformationMatrix().
|
protected |
Definition at line 88 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and v_DoMultiplybyInverseTransposedTransformationMatrix().
|
protected |
Definition at line 79 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), v_InitObject(), and v_TransformedSchurCompl().
|
protected |
Definition at line 80 of file PreconditionerLowEnergy.h.
Referenced by CreateMultiplicityMap(), SetupBlockTransformationMatrix(), v_BuildPreconditioner(), v_DoMultiplybyInverseTransformationMatrix(), v_DoMultiplybyInverseTransposedTransformationMatrix(), v_DoPreconditioner(), v_DoTransformFromLowEnergy(), v_DoTransformToLowEnergy(), and v_InitObject().
Definition at line 105 of file PreconditionerLowEnergy.h.
Referenced by CreateMultiplicityMap(), v_DoMultiplybyInverseTransformationMatrix(), v_DoTransformFromLowEnergy(), and v_DoTransformToLowEnergy().
|
protected |
Definition at line 107 of file PreconditionerLowEnergy.h.
Referenced by v_DoMultiplybyInverseTransformationMatrix(), v_DoMultiplybyInverseTransposedTransformationMatrix(), v_DoTransformFromLowEnergy(), and v_DoTransformToLowEnergy().
Definition at line 106 of file PreconditionerLowEnergy.h.
Referenced by CreateMultiplicityMap(), and v_DoMultiplybyInverseTransposedTransformationMatrix().
|
protected |
Definition at line 85 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), v_BuildPreconditioner(), and v_DoTransformToLowEnergy().
|
protected |
Definition at line 95 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), and v_TransformedSchurCompl().
|
protected |
Definition at line 97 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
protected |
Definition at line 102 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
protected |
Definition at line 92 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
protected |
Definition at line 100 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), and v_TransformedSchurCompl().
|
protected |
Definition at line 86 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), v_BuildPreconditioner(), and v_DoTransformFromLowEnergy().
|
protected |
Definition at line 90 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), and v_TransformedSchurCompl().
|
protected |
Definition at line 96 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), and v_TransformedSchurCompl().
|
protected |
Definition at line 98 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
protected |
Definition at line 103 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
protected |
Definition at line 93 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), and SetUpReferenceElements().
|
protected |
Definition at line 101 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), and v_TransformedSchurCompl().
|
protected |
Definition at line 91 of file PreconditionerLowEnergy.h.
Referenced by SetupBlockTransformationMatrix(), SetUpReferenceElements(), v_BuildPreconditioner(), and v_TransformedSchurCompl().
 1.8.1.2
1.8.1.2